Calculating ion density around a molecule
You want to calculate the ion density around a biomolecule (protein or nucleic acid)
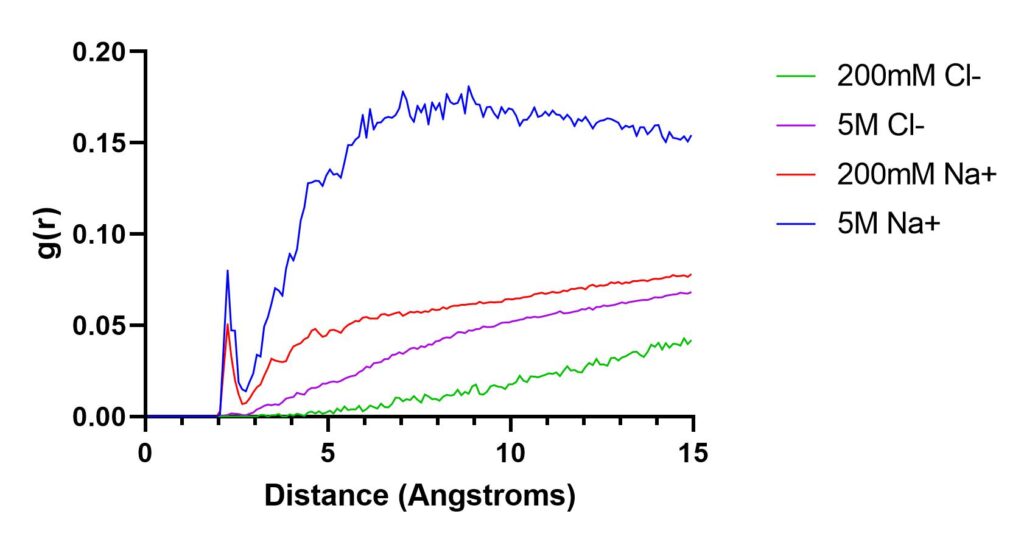
For this recipe, we can use the Radial Distribution Function available in CPPTRAJ to calculate how the density of ions varies as a function of distance from a selected reference. Our test system will be a 12-mer double stranded DNA with two different concentrations of NaCl ions, one was simulated at 200 mM and another with 5 M. With this example we will be able to see the difference of the presence of ions using the RDF command.
The trajectories of both 200mM and 5M DNA is presented in the video:
The RDF is calculated from the histogram of the number of solvent particles (mask1) to the solute (mask2). In our case, the solute is the DNA molecule (residues 1 to 24) and we have two solvent particles, sodium and chloride. The CPPTRAJ input is (for the 200mM simulation):
parm nowater.parm7 trajin nowater.nc 1 last 10 radial out 200mM-Na.dat 0.1 15.0 :1-24 :Na+ radial out 200mM-Cl.dat 0.1 15.0 :1-24 :Cl- go
The 0.1 value in the radial command is required which sets the spacing of the bins and the 15.0 value sets the maximum bin value (also required). Plotting the data files we obtain: