Create an ensemble of PDBs from a trajectory
You want to create an ensemble of PDBs from a NetCDF trajectory file.
Sometimes is useful or required to extract multiple PDB files from a trajectory. In this recipe we will use the trajout command to build a PDB file that has 10 frames from a trajectory separated by the termination card ‘TER’. This will create one single PDB file, with 10 different structures extracted from the first 10 frames of the trajectory.
For our example we will use a raw trajectory data of a molecular dynamics simulation of the crystal structure of COVID-19 main protease (PDB ID: 6LU7). We have extracted the inhibitor so only the protein is present. The fully solvated with explicit waters and ions topology file is rec.topo and a 100 NetCDF type of trajectory file is trajectory.nc. Since this is a production simulation, we need to make some pre-processing first, which involves running the autoimage command to re-orient the protein, run the rms fit command, strip the waters and the ions. After these steps, with the trajout command we set the format of the output trajectory as PDB (for more information about the formats available in the trajout comand, see this list). Then we type run to actually run the command.
Including these commands in a text file (for example: ensemble-pdb.cpptraj) you can run your analysis using:
cpptraj -i ensemble-pdb.cpptraj
The file ensemble-pdb.cpptraj looks like:
parm rec.topo trajin trajectory.nc 1 10 1 autoimage rms fit :1-306 strip :WAT strip :Na+,Cl- trajout ensemble.pdb pdb run
The parm rec.topo reads the topology, then the trajin command will read frames 1 to 10 with an offset of 1, this means that we will only read the first 10 frames of our trajectory and those frames are the ones included in the PDB file. The rms fit :1-306 will rmsd fit from residues 1 to 306 which is the size of our system, the strip command will delete any residues with the name WAT, Na+ and Cl-. Finally, we specify PDB as the file format for the trajout command. This will save the 10 frames we read with the trajin command in a PDB type of file with the name ensemble.pdb (which is available here).
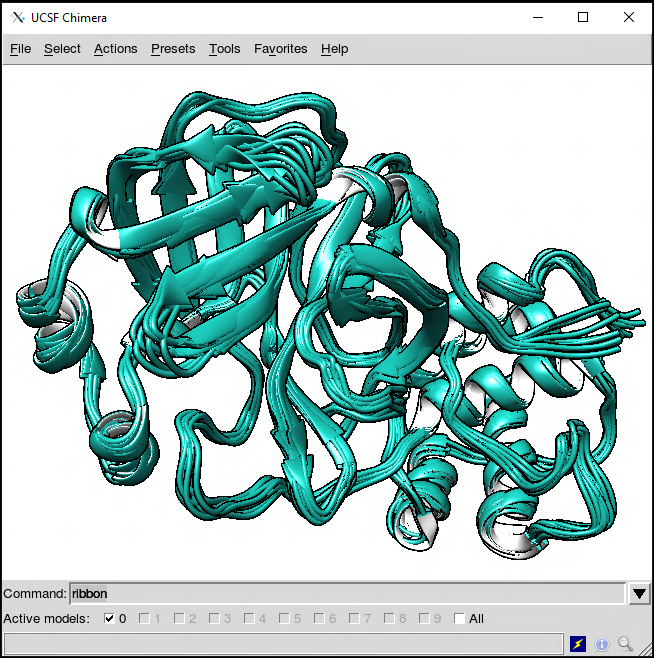
Opening the file in, for example, UCSF/Chimera, we get the 10 frames extracted by CPPTRAJ.